Last update: 17 December 2022
Introduction
Personalised medicine (PM) is a medical model that proposes to customise medical decisions, practices, and treatments for the individual patient’s specifics. It uses targeted medicines aimed at specific molecules that are involved in the patient’s disease and takes genetic, clinical, environmental, and lifestyle information about the patient into account. The aim is to select appropriate therapies for the individual patient to ensure the best possible outcome and reduce the risk of side effects.
Progress in understanding the link between genomics (and other molecular factors) and disease is an important part of the development of personalised medicine. Pharmaceutical companies are already developing some targeted medicines as a result.
The concept of Personalised medicine – The right medicine at the right dose for the right patient
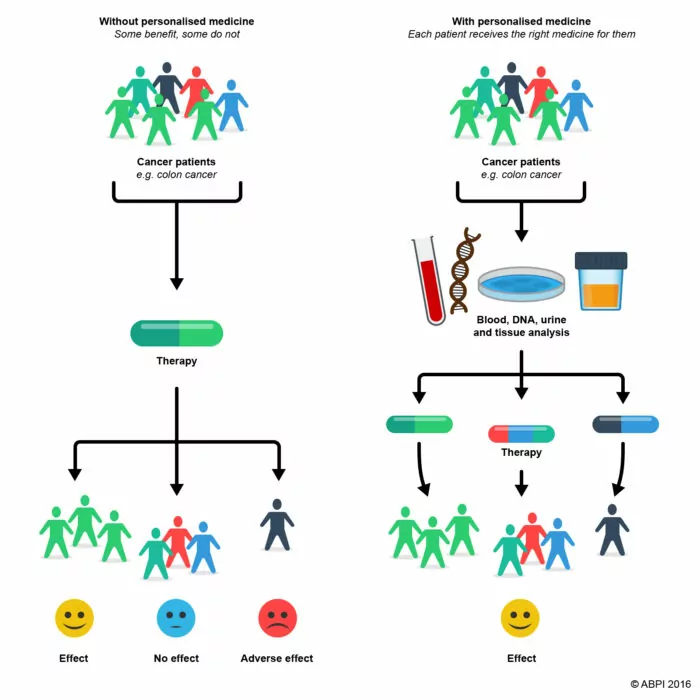
Personalised medicine is based on a detailed profile of the individual, including the sub-population that this individual patient belongs to. A doctor giving personalised medicine will be able to use targeted treatments but will consider more than just which sub-population the patient belongs to. Personalised medicine takes other information into account, such as the individual’s lifestyle and environment e.g., diet, smoking, stress. This should help to make the best possible decisions about managing the patient’s disease. In addition, the decision should be made jointly by the doctor and the patient.
Another way of adding more detail to an individual’s profile are the so-called omics technologies, the full analysis of a person’s DNA is called ‘whole genome sequencing’, this differs from the testing for variations in just one or a few genes. The whole genome sequencing technique is not a standard clinical technique as of yet. In order to better understand difference between individuals, genomic data need to be integrated with other patient information, such as their DNA structure and chemical modifications (epigenome), their proteins profiling (proteome), their metabolites profiling (metabolome) as well as comprehensive details on diet, lifestyle and environment.
Combined data from whole genome sequencing and other related technologies will help drive forward truly personalised medicine.
- Figure 1. Example schematic of use of personalised medicine. From ABPI 2016
Stratified and personalised medicines
Same symptoms, same disease, same treatment?
It has generally been thought that patients diagnosed with the same disease have the same root cause and as such, were offered the same treatments. However, clinical experience shows that not all patients respond in the same way to the same treatment.
In reality, patients diagnosed with the same disease can have different causes for their disease and experience different symptoms. Therefore, it is important to develop medicines or treatments tailored towards a specific (sub-)group of patients or individuals who manifest the same symptoms.
Biomarker profiles are fundamental for identifying patients that share similar characteristics and allowing patient stratification in subgroups.
Personalised Medicine [PM]
The European Alliance for Personalised Medicine (EAPM) defines personalised medicine (PM) as ‘a targeted approach to the prevention, diagnosis and treatment of disease based on an individual’s specific profile’. The main aim of (PM) is to implement/use measures/technologies to treat the right patient (meaning correctly diagnosed) with the right medicine at the right time.
An individual’s health is determined by their genetic differences combined with their lifestyles and environment. By combining and analysing patient characteristics, patterns can be identified that can help to determine an individual’s risk of developing disease; detect illness earlier; and, determine the most effective interventions to help improve health, be they medicines, lifestyle choices, or changes in diet.
Progress in understanding the link between genomics (and other molecular factors) and disease is an important part of the development of personalised medicine.
- Figure 2. European Alliance of Personalised Medicine
Stratified Medicine
Stratification, in the present context, means defining sub-populations (a group or proportion of patients) according to which disease ‘sub-type’ an individual has been diagnosed with. Stratification helps identifying patients with distinct mechanisms of disease, or the likelihood of particular responses to treatments, allowing to identify and develop treatments that are effective for particular groups of patients.
Breast cancer can be linked with hormones, e.g. oestrogen and progesterone. Most breast cancer cells, but not all, have a large number of molecules (receptors) that bind with oestrogen and allow these cells to grow when oestrogen is present – these cells are ‘ER positive’. Many of these cells also grow in response to progesterone – they are ‘PR positive’ cells. ER- and PR-positive cells are called “hormone-receptor positive cells” (Travis, C. R., Key, T. J.:2003).
Breast cancer cells that are ER and/or PR positive are likely to respond to medicines that block the action of oestrogen or progesterone. Around 60 in every 100 breast cancer patients respond to these medicines. The same medicines will only be effective for around 5 to 10 breast cancer patients in every 100 if the tumour is not ER and/or PR positive.
Some breast cancer cells also produce too much of a protein called ‘Her2/neu’. They are known as ‘HER-2 positive’ and tend to be more aggressive as the cells tend to grow much faster. The medicine trastuzumab binds to the Her2/neu protein and stops the cancer cells from growing and dividing, resulting in an improvement of the overall survival for HER-2 positive patients with breast cancer[1].
Some breast cancers are neither ER, PR, nor HER-2 positive. These tumours are called ‘triple negative’, and as yet, no targeted therapies are available for these types of tumours. Therefore, more usual types of chemotherapy will be prescribed for these patients.
Stratified medicine is well developed in the cancer field, but there are many gaps in other diseases (e.g., neurological diseases), especially if the disease cause is multifactorial, and not only genetic.
Often the terms ‘personalised medicine’ and ‘stratified medicine’ are used interchangeably. The table below showcases the key differences between these 2 terms:
| Personalised medicine | Stratified medicine |
|---|---|
| Aims to use measures to treat the right patient with the right dose at the right time. PM uses targeted medicines taking into account other individual information to tailor the treatment and management of the patient to their particular situation. It is used to ensure the best outcome and reduce the risk of side effects. | It is the use of a medicine that is targeted at a patient sub-population (a group or a proportion of patients, e.g. having a particular disease, age group or disease stage), instead of using one medicine to treat all patients with that disease |
Table 1: Stratified and personalised medicines. EUPATI Toolbox.
Personalised and stratified medicines are growing in importance, and their approaches have been widely applied to cancer treatments and rare diseases, where there is plenty of available information on the underlying genetic causes of the disease (Verma:2012). It is expected that these approaches will be increasingly used in the development of treatments for other diseases as well. This is an important change in the approach to treating patients based on the underlying cause of their specific disease. Although people may be diagnosed with the same disease, clinical experience shows that people may respond differently to the same treatment:
- some will respond well
- some will not respond, but not to the same level as others
- some will not respond at all
- some may have serious side effects (adverse reactions)
- some may develop ‘resistance’ to the medicine (they stop responding) - this can happen even if they responded well at first.
When treating a patient, doctors try to take into account various clinical and lifestyle factors for individual patients. However, often the only option is to try a treatment, monitor the patient’s response, and change the dose, schedule or medicine if necessary. It is now known the diseases of these patients are due to different causes. In other words, the same symptoms shown by the patients can be caused by different processes happening in their cells (they have a different underlying pathology, different genomic and genetic factors etc.). These differences can have a great impact on the way a disease progresses, and how an individual responds to a specific treatment.
Biomarkers
Processes that happen at the level of cells and molecules can be measured using ‘biomarkers’. Biomarkers are essential tools to enable PM, for example, the selection of appropriate patients for treatment with tailored medicines.
The word biomarkers here refers to a biological marker, something that can be measured and points to the presence of a disease, a physiological change, a response to a treatment, or a psychological condition.
For example, glucose levels are used as a biomarker in managing diabetes, and brain images can provide information about the progression of multiple sclerosis[2].
Biomarkers are used in many scientific fields. They are used in different ways at different stages of medicines development, including in some cases as a surrogate endpoint to indicate and measure the effect of interventions, such as medicines, in clinical trials.
For example, haemoglobin levels have been used in Phase III trials to support development of therapies for Type 1 Gaucher disease (a rare disease that affects multiple organ systems and shortens life expectancy, but which can take years to show changes in clinical symptoms)[3].
Biomarkers used for PM are a part of a relatively new clinical toolset. Complex biomarkers signatures might be the result of multi-omics data integration.
They are categorised in different ways according to their clinical applications. Biomarkers have a clinical role in narrowing or guiding treatment decisions. Some examples of biomarkers are:
- Physiological measures such as blood pressure or temperature.
- Biological substances (‘biochemicals’), such as enzymes or hormones.
- Gene changes.
- Images from Magnetic Resonance Imaging (MRI).
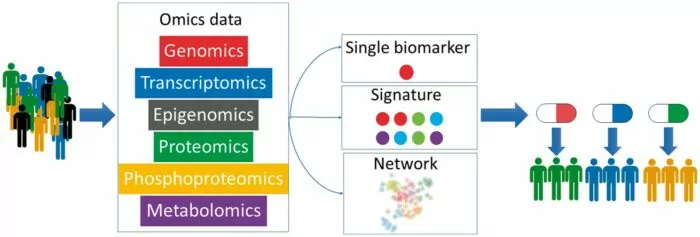
Multi-omics technologies
Multi-omics technologies are used to analyse biological processes that happen at different levels, such as at the DNA (genome) and DNA modification (epigenome) level, RNA (transcriptome) level, protein (proteome) and metabolites (metabolome) level.
By combining these, scientists can analyse complex biological data to better understand diseases and identify biomarkers.
Current research is telling us a lot about diseases from a cell and molecular level. There is evidence that specific changes to genes (variations) can affect the way cells function and how a disease may develop. Diseases with similar symptoms may be diagnosed as the same disease. However, they may be caused by different genetic variations.
Transcriptomics
Transcriptomics is the study of the transcriptome - the complete set of RNA transcripts that are produced by the genome, under specific circumstances or in a specific cell - using high-throughput methods, such as microarray analysis. Comparison of transcriptomes allows the identification of genes that are differentially expressed in distinct cell populations, or in response to different treatments.
This growing area helps us to understand variation in disease. Twin studies revealed how people with the same genomic background can have different risk for a certain disease. This can be explained by epigenetics. Epigenetics is the study of how genes can be turned on or off or modulated (turned up or down) by environmental factors, without changes to their DNA sequence. Epigenetic changes may influence how a patient responds to treatment, since the changes can occur in response to environmental or lifestyle factors, such as UV light exposure, diet, smoking, or stress. Ultimately, a personalised medicine would take account of an individual’s epigenetics.
Proteomics is the large-scale study of proteomes. A proteome is a set of proteins produced in an organism, system, or biological context, for instance, the proteome of a species (for example, Homo sapiens) or an organ (for example, the liver). The proteome is not constant; it differs from cell to cell and changes over time. To some degree, the proteome reflects the underlying transcriptome. However, protein activity is also modulated by many factors in addition to the expression level of the relevant gene.
Proteomics is used to investigate:
- when and where proteins are expressed,
- rates of protein production, degradation, and steady-state levels,
- how proteins are modified,
- the movement of proteins between subcellular compartments,
- the involvement of proteins in metabolic pathways,
- how proteins interact with one another.
Proteomics can provide significant biological information for many biological problems, such as:
- which proteins interact with a particular protein of interest,
- which proteins are localised to a subcellular compartment,
- which proteins are involved in a biological process.
Metabolomics
Metabolomics is the large-scale study of small molecules, commonly known as metabolites, within cells, biofluids, tissues or organisms. Collectively, these small molecules and their interactions within a biological system are known as the metabolome.
Just as genomics is the study of DNA and genetic information within a cell, and transcriptomics is the study of RNA and differences in mRNA expression; metabolomics is the study of substrates and products of metabolism, which are influenced by both genetic and environmental factors. Metabolomics is a powerful approach because metabolites and their concentrations, unlike other “omics” measures, directly reflect the underlying biochemical activity and state of cells/tissues.
Pharmacogenetics or pharmacogenomics
One type of biomarker that is becoming widely used is an individual’s genetic or genomic information. The study of how genetics and genomics affect an individual’s response to a treatment is known as pharmacogenetics and pharmacogenomics. These help to “tailor a treatment” according to an individual’s genetic configuration. Few medicines on the market today require a genetic test before the medicine is prescribed, to ensure the treatment is safe for the individual patient. For example:
HIV patients are tested for a genetic variation known as ‘HLA B*5701’ before the medicine abacavir is prescribed because the variation is associated with an adverse reaction to the medicine.
In addition, the information provided with some other medicines on the market advises doctors how to use a patient’s genomics information when they prescribe the medicine. Genomics information can help to decide if that medicine is the best option for that patient, and what would be the best dose.
For example:
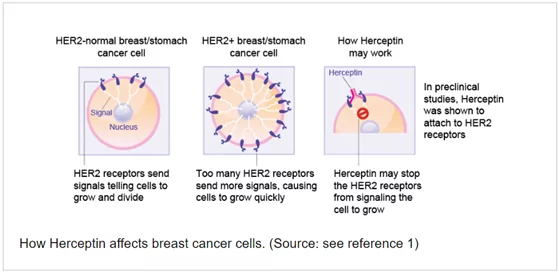
Trastuzumab (Herceptin) is a gene-targeted monoclonal antibody directed against the Human Epidermal growth factor Receptor-2 (HER-2) and approved for the treatment of breast cancer. HER-2 is overexpressed in approximately 20% of breast cancers, causing an excessive signalling to the cell interior that drives breast cancer cells to grow faster and faster. HER-2 exists on the surface of some breast cancer cells and is routed in the cell membrane. Only patients who have tested positive for high HER-2 levels will benefit from treatment with trastuzumab. Trastuzumab works by attaching itself to the HER-2 on the surface of breast cancer cells and blocking them from receiving growth signals. By blocking the signals, trastuzumab can slow or stop the growth of the breast cancer and is an example of an immune targeted therapy.
- Figure 3. How Herceptin affects breast cancer cells
Biomarkers and medicines development
To create targeted treatments, biomarkers are increasingly used in medicines development. This is expected to:
- improve outcomes for patients: doctors should be able to choose medicines that work well for each individual, and with a lower risk of serious side effects,
- improve the efficiency of medicines development, making non-clinical and clinical trials more efficient, less time consuming and safer.
After identifying a biomarker for a specific condition, must go through the experimental and analytical validation. Experimental validation allows the development of the most adapted protocol for routine use of the biomarker. Analytical validation is one of the most important steps, it serves to identify specific characteristics of the candidate biomarker before developing a routine test. Several parameters are considered, including sensitivity, specificity, robustness, accuracy, reproducibility etc. Then, the protocol standardisation optimises the validated protocol for routine use, including analysis of the critical points by scanning the entire procedure to identify and control the potential risks.
- Figure 4. Proteomics and phosphoproteomics in precision medicine applications and challenges
Medicines development and Personalised Medicine
Personalised medicine and industry
Pharmaceutical companies have already begun producing targeted medicines. Many have committed to the idea of tailored treatment, particularly for oncology. However, PM presents both opportunities and challenges, allowing for substantial clinical advances and accelerated medicine development timelines. Costs may be increased due to the complexities of biomarker analysis and diagnostic development. Biomarkers need to be validated and reliable, requiring new competencies in trial design, data analysis and investigator expertise in sample collection and management. Moreover, greater statistical and computing power is necessary to mine data to better understand the relationships between genetic, biological and environmental factors. Because of this, pharmaceutical industry, and relevant stakeholders – academics, regulatory agencies, payers and policy makers - should be committed to working together.
Companion Diagnostics
A Companion diagnostic is a medical device, often an in vitro device, which provides information that is essential for the safe and effective use of a corresponding medicine or biological product. They are necessary tests that help select patients before a medicine is given. They may:
- show who is likely to respond to the medicine (‘responders’ and ‘non-responders’),
- identify patients at high risk for adverse reactions,
- help the doctor to select an appropriate dose that is both safe and effective.
Personalised approaches translation in clinical setting: the importance of companion diagnostics
Increasingly, new medicines are authorised together with an accompanying diagnostic test to ensure that the medicine is appropriate for the patient to be treated. If a diagnostic test is not properly validated, it won’t be able to identify the right patient that could benefit from this medicine or patient may experience side effects due to being treated with suboptimal medicine. Therefore, proper design and validation of the diagnostic test is essential to obtain the best outcome for the patient from the treatment.
Companion diagnostics may include tests directly on the patient such as electrocardiography (ECG) or diagnostic imaging such as MRI. Tests carried out on samples taken from a patient (such as DNA tests) are generally considered by regulatory authorities to provide the most valid evidence.
Companion diagnostics should have an ‘intended use’ or ‘indications for use’. They are often both referred to under the umbrella term of ‘intended use’ (of a treatment). They should generally include the following elements:
- The target population for whom the test is intended (such as individuals with particular genotypes (genetic make-up) or phenotypes (characteristics)).
- Why the measurement is being done – the ‘clinical purpose’ (such as to help with diagnosis, assessing how the disease is likely to develop (prognosis), and monitoring).
- What is measured, identified, or detected (such as a specific gene or protein).
- What kind of measurement the test makes, including whether the test is qualitative (looks at observations and descriptions), or semi-quantitative or quantitative (looks at numbers).
- The sample type and where it is taken from (e.g., whole blood, cerebrospinal fluid).
- The setting in which the diagnostic device is meant to be used (in a laboratory or at ‘point-of-care’) and what type of equipment is required to perform the test.
- The target condition (a particular disease, disease stage, health status, or any other identifiable condition or event).
Challenges and opportunities in personalised medicine
Treating patients with personalised medicine will require the development of targeted medicines.
For personalised medicine to progress, new findings from molecular research, and new technologies (such as ‘omics’ technologies), must be translated (adapted) for use in medicines development and approved therapy.
However, an advantage of developing targeted medicines is the increase in the efficiency of clinical trials. Fewer new medicines should fail at each stage of the development process if they are targeted at a known cause of the disease, and should be combined with appropriate biomarkers. Validation of biomarkers as unique and predictive tools for the outcome of treatment should be in place before medicines developed in this way can be used more widely.
Biobanks
Studies that use biobanks are especially important for the development of personalised medicine, and biobanks are increasingly used in clinical trials for new medicines. Biobanks are basically large, organised sets of blood and/or tissue samples donated in line with applicable legislation by patients and healthy volunteers. They also include carefully collected data on the donors’ clinical condition, lifestyle (diet, smoking, etc.) and other factors. Biobanks allow the cells and molecules of large numbers of samples to be studied, and for this information to be linked with clinical and other data. Combining information in this way is helping to understand why individuals vary:
- in which diseases they develop,
- in how severe their diseases are, and,
- in how they may respond to treatment.
The more samples available, the more effective such studies can be.
Biobanks are being set up in many countries. BBMRI-ERIC is a European research infrastructure for biobanking. They bring together all the main players from the biobanking field – researchers, biobankers, industry, and patients – to boost biomedical research. To that end, they offer quality management services, support with ethical, legal and societal issues, and a number of online tools and software solutions. Another biobank network is ‘EuroBioBank’, that links biobanks from different countries to make even more data available for research (in this case, into rare diseases).
Personalised approaches translation in clinical setting: the challenges of patients and doctors
Ethical Challenges
There are particular ethical challenges for researchers who work with personal data. It is important that they consider privacy and keeping data confidential (see GDPR) and making sure to obtain written consent. See also EUPATI Open Classroom, Module 3, ‘Non-clinical development’ Course 3 Book - Limitations, challenges and ethics.
Patient experience
Personalised medicine starts with the patient. Indeed, PM can provide much more information about the health of an individual person, which can have personal implications and potentially change their way of living. It is, however, a complex discipline, and if patients are to make informed decisions about health-related issues, it will be vital for them to have the necessary knowledge. In other words, patients need to be 'empowered' to become participants in their own healthcare. Are patients ready for this? And are healthcare professionals skilled enough and ready to communicate about the implications of PM with their patients?
With personalised medicine, the patient with a disease should have more reassuring information prior to treatment:
- that a medicine is more likely to work for them and why, and
- that they are less prone to experience side effects with a particular treatment.
Where side effects are unavoidable, better knowledge about them and how severe they might be should make it easier for the patient to decide and plan for the treatment and to fit it into their daily life.
Patient-doctor communication
In general, with personalised medicine, there may exist more or different information about the available treatment options for the patient and doctor to understand and discuss. Patients not familiar with PM may need additional support from their physician to ensure they understand the information and can make an informed decision.
It may be necessary to do more tests than generally applied in order to enable personalised medicine. Blood tests are generally seen as routine, but biopsies (where samples of tissue are removed) require an anaesthetic and can take longer to be analysed. Patients and their doctors will need to discuss the pros and cons when making decisions about testing. This discussion will help to take the appropriate decision for the patient.
If a test predicts how likely a patient is to respond to a medicine, the result might be expressed for example as ‘odds’ (1 in 3) or as percentage (33 %). People have different ways of interpreting risks and benefits, and physicians will need to provide support to ensure an appropriate interpretation of risks and benefits is being done by the patient.
Further resources
- Genomics England
- Nuffield Council on Bioethics (2010). Medical profiling and online medicine: the ethics of ‘personalised healthcare’ in a consumer age.
- Corpet, A. & Almouzni, G. (Dec 2006-Jan 2007).Sciences et Avenir, 149
- "The Case for Personalized Medicine"(PDF). Personalized Medicine Coalition. 2014
- Smith R (15 October 2012). "Stratified, personalised, or precision medicine". British Medical Journal.
- Jackson and Chester “Personalised cancer medicine” DOI: 10.1002/ijc.28940
- EUROPEAN ALLIANCE FOR PERSONALIZED MEDICINE: https://www.euapm.eu/
Attachments
- Challenges-in-Personalised-Medicine-v1_EN
Size: 396,441 bytes, Format: .pptx
A presentation describing the challenges in personalised medicine, which can be adapted for own use.
References
- How Herceptin affects breast cancer cells” by beyondthedish.wordpress.com is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License.
See: https://beyondthedish.wordpress.com/2012/06/04/smart-bomb-successfully-treat-advanced-breast-cancer-in-clinical-trials/ - Travis, C. R., Key, T. J. 2003. Oestrogen exposure and breast cancer risk. See: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC314432/
- Verma, M. 2012. Personalized Medicine and Cancer. See: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4251363/
Footnotes
1 Breast Cancer HER2 Status. www.cancer.org